Introduction
Traumatic Brain Injury (TBI) is a serious concern for both the military and the general civilian population. In particular, blast-related TBI has been prevalent in recent military conflicts [1]. During Operation Iraqi Freedom, a study of casualties requiring level V care at Walter Reed Army Medical Center reported that 29% of those screened had a TBI. Blast and explosion were the most common cause, accounting for 78% of those found to have a TBI [2]. Likewise, in the civilian population, approximately 1.4 million people in the United States sustain a TBI each year. Of that number, 50,000 die, 235,000 are hospitalized, and 1.1 million are evaluated, treated, and released from the nation’s emergency departments [3]. When one also includes concussions (often called mild TBIs) in the discussion, it is also possible that the largest proportion of patients is never even seen in an emergency department. But perhaps the most concerning aspects of TBI are the residual effects, with at least 5.3 million Americans—almost 2% of the population—estimated to have current long-term or lifelong disabilities from TBIs [4].
TBI associated with closed head injuries, also referred to as nonpenetrating head injuries, can be caused by blast, blunt force impact, or sudden acceleration. In cases such as these, diffuse axonal injury is one particular injury mechanism that has been cited as a signature injury of TBI neural damage [1, 5]. Deformation of the brain tissue can induce misalignment in the cytoskeletal network or axolemma permeability, inducing a cascade of subcellular events and culminating in the severance of the axon [6, 7]. It is these axon fiber bundles that make up the structural network that allows neurons to communicate with one another. Injury to the axons leads to degraded structural connectivity, which may be responsible for the cognitive deficits that are characteristic of mild, moderate, and severe cases of TBI [8]. Concussion, or mild TBI, is thought to be a less severe type of diffuse axonal injury, where axons are damaged to a minor extent from stretching. Postmortem studies of brains with concussions have found axonal damage; however, because of other factors, such as restricted blood flow, it is not possible to isolate the cause of this damage and solely link it to concussions.
Numerical Approach
Finite element (FE) simulations are often used to better understand the mechanical response of the brain and investigate the potential for neurotrauma during a blast or impact event. Magnetic resonance imaging (MRI) scans can provide valuable detail on the geometry of the head, skull, and brain. The data from these scans can be used to create accurate, patient-specific FE meshes that can then be used in these blast or impact simulations. Tissue damage is computed using empirically based damage models. Modeling axonal injury mechanisms within the white matter of the brain has been the focus of many recent efforts. Various biomechanical and physiological injury thresholds for neurotrauma have been proposed, including those based on intracranial pressure [9] and those based on axonal strain [10, 11, 12]. Simulations using strain-based injury criteria have shown that the degree of injury predicted is highly dependent on the incorporation of the axonal orientation information and the inclusion of material anisotropy into the constitutive model for white matter.
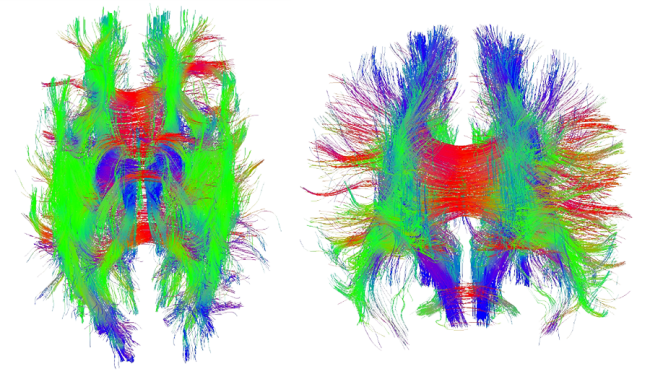
Traditional MRI scans are not able to provide the requisite detail at the microstructural level to generate the orientation vectors needed in the anisotropic material models. To obtain data on the axonal fiber orientation, a relatively new noninvasive tool called Diffusion Tensor Magnetic Resonance Medical Imaging (DT-MRI)—or Diffusion Tensor Imaging (DTI)—is used to capture the structural organization of the white matter. DTI is based on the principle that within biological tissue, water diffuses more rapidly in the direction aligned with the internal structure and slower in the perpendicular directions. The resulting images can be used to generate the diffusion tensor, from which diffusion anisotropy measures, such as the fractional anisotropy, can be computed. The principal direction of the diffusion tensor can be used to estimate the white matter connectivity of the brain. As shown in Figure 1, DTI allows the axonal fiber tracts to be charted and then imported into an FE model so that they can influence the mechanical response of the brain. These models can then be used to calculate the strains and damage in these fibers to create a prediction of diffuse axonal injury [10, 13].
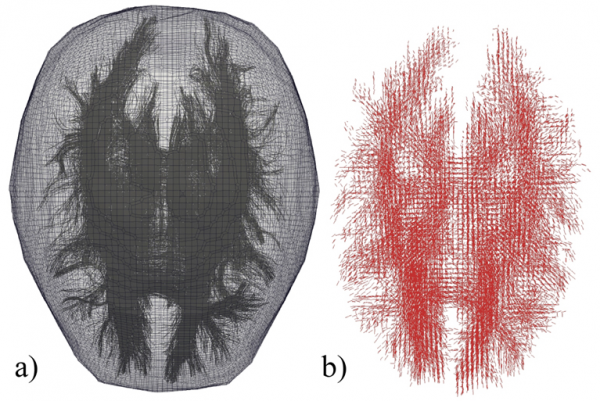
The anisotropic material model, which is described in the next section, requires a unit vector for each element to identify the fiber orientation, ![]() . This task is complicated due to the unstructured nature of the FE mesh and the differences between the DTI scan resolution and that of the FE mesh. Fibers may begin and end in different elements, and a single element may contain multiple fibers. An algorithm is used to take the DTI data and assign an orientation vector to each element within the FE mesh. To begin, each fiber is subdivided into a number of smaller straight-line segments. The algorithm determines if any fiber segment overlaps with a given FE. If no fibers overlap an element, then that element is treated as an isotropic material. Depending on the fiber density and the element size there may be multiple fibers or multiple fiber segments within a single element. In this case, an averaging scheme is used to assign a single orientation to that element. The scheme adds all of the fiber segment vectors (from the same fiber) that are contained within the element. The resulting vector is normalized to create an average orientation. Note that the fiber segment vectors are not unit vectors, so their physical length acts as a weighting function in the averaging scheme. If multiple fibers pass through the element, the process is repeated for each fiber, after which all of the resulting averages are summed and normalized. There is the potential to include multiple fiber families, each with their own orientation, by expanding upon the following material model. This expansion would be of the greatest potential use when combined with Diffusion Spectrum Imaging (DSI), which is capable of describing fiber crossings. An example of the discretized axon fiber tracts along with the FE mesh is shown in Figure 2.
. This task is complicated due to the unstructured nature of the FE mesh and the differences between the DTI scan resolution and that of the FE mesh. Fibers may begin and end in different elements, and a single element may contain multiple fibers. An algorithm is used to take the DTI data and assign an orientation vector to each element within the FE mesh. To begin, each fiber is subdivided into a number of smaller straight-line segments. The algorithm determines if any fiber segment overlaps with a given FE. If no fibers overlap an element, then that element is treated as an isotropic material. Depending on the fiber density and the element size there may be multiple fibers or multiple fiber segments within a single element. In this case, an averaging scheme is used to assign a single orientation to that element. The scheme adds all of the fiber segment vectors (from the same fiber) that are contained within the element. The resulting vector is normalized to create an average orientation. Note that the fiber segment vectors are not unit vectors, so their physical length acts as a weighting function in the averaging scheme. If multiple fibers pass through the element, the process is repeated for each fiber, after which all of the resulting averages are summed and normalized. There is the potential to include multiple fiber families, each with their own orientation, by expanding upon the following material model. This expansion would be of the greatest potential use when combined with Diffusion Spectrum Imaging (DSI), which is capable of describing fiber crossings. An example of the discretized axon fiber tracts along with the FE mesh is shown in Figure 2.
Figure 2. DTI Data Overlaid Onto a Brain’s FE Mesh (a) and Discretized Orientation Data Assigned to Each Mesh Element (b).
With an anatomically accurate mesh of the head, skull, and brain, along with constitutive equations informed by diffusion-weighted imaging, FE simulations of blast and impact can be carried out. These simulations produce a spatio-temporal description of the tissue deformation. The resulting strains and pressures can be compared to experimentally based injury thresholds for white matter, relating how the simulated blast or blunt impact forces translate to cellular death in neural tissue. Using high-performance computing clusters, we are then able to probe these forces at spatial scales matching our neuroimaging efforts. However, reliable predictions of tissue deformation are highly dependent on accurate constitutive equations describing the stress-strain response of the various biological materials.
We now consider one of the material models that will be used to describe the white matter and its axon fiber bundles.
Transversely Isotropic Material Model
In many biological tissues, fibers or bundles of cells are aligned in uniform directions. As a result, isotropic materials models are insufficient for capturing the mechanical behavior. For the white matter within the brain, axon bundles form complex fiber tracts as they connect and facilitate communicate between different regions in the brain. These axonal fiber tracts have been reported to be approximately three times stiffer than the surrounding matrix material [14] and thus play an important role in the mechanical response of the brain. To model the white matter, we use a transversely isotropic hyperelastic material, where the fiber tract directions are determined from DTI data. A viscoelastic material model can be employed to include rate effects; however, for the sake of simplicity, rate effects are not discussed here.
To describe the material model, we must first define some basic kinematic concepts. The deformation gradient is defined as:
![]() ,
,
where ![]() is the position of a material point in the reference (undeformed) configuration and
is the position of a material point in the reference (undeformed) configuration and ![]() is the position of the same material point in the current (deformed) configuration. The ratio of the deformed volume to the undeformed volume is given by the Jacobian, the determinant of the deformation gradient:
is the position of the same material point in the current (deformed) configuration. The ratio of the deformed volume to the undeformed volume is given by the Jacobian, the determinant of the deformation gradient:
![]() .
.
It is often beneficial to perform a multiplicative decomposition of ![]() into volume-changing (dilational) and volume-preserving (distortional) parts to separate the bulk and the shear response. To accomplish this decomposition, a deviatoric deformation gradient in which the volume change is eliminated is defined as:
into volume-changing (dilational) and volume-preserving (distortional) parts to separate the bulk and the shear response. To accomplish this decomposition, a deviatoric deformation gradient in which the volume change is eliminated is defined as:
![]() .
.
We can then define a modified right Cauchy-Green tensor:
![]() .
.
The modified principle invariants of the right Cauchy-Green deformation tensor are defined as:
![]() ,
,
![]() ,
,
and
![]() .
.
For the modified right Cauchy-Green tensor, because the volume change has been eliminated, the third principle invariant will always be 1. To capture the anisotropic nature of the white matter, we introduce a unit vector, ![]() , assigned using DTI data, that describes the direction of the fiber in the undeformed reference configuration. We can then define two additional invariants based on the fiber direction:
, assigned using DTI data, that describes the direction of the fiber in the undeformed reference configuration. We can then define two additional invariants based on the fiber direction:
![]() ,
,
and
![]() ,
,
where ![]() and
and ![]() arise from the anisotropy and describe the deformation of the fiber family. Should it be necessary to include multiple fiber directions, we can define additional invariants for each new fiber family(
arise from the anisotropy and describe the deformation of the fiber family. Should it be necessary to include multiple fiber directions, we can define additional invariants for each new fiber family(![]() , and so on). It should be noted that
, and so on). It should be noted that
![]() ,
,
where ![]() is the direction of the fiber in the current configuration and λ is the stretch in the fiber bundle. Thus,
is the direction of the fiber in the current configuration and λ is the stretch in the fiber bundle. Thus, ![]() will also be useful in evaluating strain-based injury criteria for the axon fiber bundles.
will also be useful in evaluating strain-based injury criteria for the axon fiber bundles.
The strain energy, Ψ, of the transversely isotropic hyperelastic material can be written as a function of the modified principle invariants along with the Jacobian, which describes the change in volume. Assuming that the responses of the fibers and the matrix material are not strongly coupled, we can choose to separate the strain energy into a linear combination of the isotropic and anisotropic components [13, 15, 16]:
![]() ,
,
where ![]() describes the response of the isotropic matrix and
describes the response of the isotropic matrix and ![]() describes the directional contribution of the reinforcing fiber bundles. We can then select an appropriate isotropic strain energy function for the matrix component, such as a Neo-Hookean or Mooney-Rivlin material model. For the anisotropic response, it is suggested to select a Fung material model that includes the exponential behavior characteristic of most soft tissues [16]:
describes the directional contribution of the reinforcing fiber bundles. We can then select an appropriate isotropic strain energy function for the matrix component, such as a Neo-Hookean or Mooney-Rivlin material model. For the anisotropic response, it is suggested to select a Fung material model that includes the exponential behavior characteristic of most soft tissues [16]:
![]() ,
,
where ![]() and
and ![]() are material constants obtained from a parameter fit to experimental data. This example is a relatively simple example of a Fung material. Depending on the available data, a more complex constitutive model for the fibers may be preferred.
are material constants obtained from a parameter fit to experimental data. This example is a relatively simple example of a Fung material. Depending on the available data, a more complex constitutive model for the fibers may be preferred.
Numerical Simulation
A 3-D finite element model of the human head can be constructed using MRI data [17, 18]. The work described here used a Lagrangian, 3-D, explicit transient dynamic code called Presto within the Sierra Solid Mechanics suite of codes from Sandia National Laboratories. While this article is focused on modeling the white matter, the full simulation requires constitutive descriptions and properties for all of the components, including the skull, cortex, brain stem, cerebrospinal fluid, and skin and muscle (usually represented together as a soft tissue mixture). Within each volume element in the white matter region of the mesh, an algorithm assigns a fiber direction based on the DTI data. The model can be exposed to a variety of loading conditions, such as an applied blast pressure or an impact, both potential causes for TBI. Within the simulation, the axonal strain is calculated and compared against an injury threshold. The literature contains a wide range of such injury thresholds, from 0.05 to 0.21 [19, 20]. Bain and Meaney [11] have shown an electrophysiological impairment at axonal strain levels of 0.28 (liberal), 0.13 (conservative), and 0.18 (optimal) in tension.
In one series of simulations, a blast pressure was applied based on a charge detonated 3 m in front of the body at mid-thoracic height. The size of the charge was chosen so as to produce a peak incident pressure of 210 kPa and a positive duration of 2.8 ms. This loading is survivable with ballistic protective body armor and falls below the 50% survivability threshold for lethal head injury [21]. The largest axonal strains produced during this loading exceeded 0.17 and increased at the center of the brain throughout the simulation. Elements on the surface of the brain and near the ventricles also exhibited large amounts of axonal strain. Peak axonal strains occurred approximately 10–15 ms after the initial contact of the blast wave. Simulations were run with the head fixed and with the head permitted to rotate. When head rotation was prohibited, axonal strains did not exceed even the conservative threshold for electrophysiological impairment. Allowing the head to rotate in response to the blast wave and the associated angular acceleration plays a significant role in the calculation of axonal strains and contributes to the severity of the TBI [22]. In further studies, frontal blast loading was compared to side blast loading. The side blast led to a higher concentration of axonal degradation in the occipital region, while frontal blast produced a more evenly distributed axonal damage [23].
Another series of simulations looked at diffuse axonal injury caused by frontal impact to the head. To mimic the impact, a time-dependent force was applied to a 3-mm2 circular area on the forehead. The force-time curve was sinusoidal in shape, with a peak force of 7,000 N at 2.7 ms. While the highest pressures were seen in the frontal region of the brain, the largest axonal strains occurred in the temporal and occipital regions. In these areas, the axonal strain exceeded the 0.18 threshold for electrophysiological impairment [18].
Conclusion
The modeling efforts discussed herein have focused on the use of noninvasive DTI techniques to, in effect, provide a signature of TBI. In addition, these efforts have potential for even greater predictive capabilities investigating the structure-function relationship within the brain and the neurological impact of the axonal damage. The brain’s gray matter contains most of the neuronal cell bodies involved in decision-making, memory, sensory perception, and muscle control; the white matter, made up of the bundles of axons, is responsible for communication between the different regions of gray matter. Using DTI, the paths of the fibers can be charted; regions of the brain associated with specific roles can be identified by subject-matter specialists or through scans (such as a Functional Magnetic Resonance Imaging [fMRI]); and the fiber tracts connecting these regions can be examined. While these simulations are not at a level of detail needed to map every connection within the brain (the current goal of the Human Connectome Project), we can still (using graph theory) look at what regions are connected and measure the strength of those connections, both through the number of fibers and the length of the connections.
A map of the neural pathways can be generated where specific regions of gray matter are represented as nodes with connections based on the axon fiber paths. Connections passing through the regions where strain is above a certain threshold, as predicted by the FE simulation, are degraded. The strength of the communication in this neural network can be calculated both before and after the simulation. Network science provides the means to calculate the global and local efficiency of these networks, as well as their capability to transfer information between nodes. By comparing these measurements both before and after the traumatic event, the reduction in the brain’s capacity for communication can be calculated, thus providing a measure of mental impairment. Accordingly, this approach provides not just a means to predict the amount of cellular damage within the brain but also to predict the cognitive effects that damage may have on an individual.
In addition, while the example simulations discussed herein have focused on wartime injury (blast and projectile impact), this approach has extensive applications for modeling TBI within the civilian population as well. As mentioned previously, concussions are a mild form of TBI that are a major concern in both amateur and professional sports. There is considerable public pressure to increase the safety of athletes, especially at the high school and college levels. Also, the accelerative loading to the head during an automobile crash is another common cause of TBI within the United States. Thus, this approach to modeling diffuse axonal injury could help in evaluating new designs for protective sports gear and automotive safety measures (as discussed in the inset article).
Not surprisingly, validation of these simulations remains a challenge. Human experiments have been limited to cadaveric testing, where pressures, displacements, and strains can be measured. These measurements, when available, have shown good agreement with the simulation; however, experimental measurements of axonal damage and cell death (the quantities of greatest interest) are not possible through cadaveric testing. Qualitative comparisons can be made from medical scans, but true validation would require scans both before and after the head trauma. The geometry of the brain and the white matter fiber tracts are patient-specific, so without pre-injury DTI data, comparison with post-injury data is approximate at best. The use of animal testing as a human analog is one possible alternative; however, there are ethical implications of animal testing which must also be taken into consideration. Hopefully, long-term comprehensive studies of both soldiers and athletes will be published and will provide repeated DTI scans to investigate changes in the white matter tracts over time. For example, pre- and post-game DTI scans of football players could be combined with in-helmet accelerometers to record loading and identify axonal injury. Studies such as these have the potential to provide further insight to fine tune and validate the numerical models.
References:
- Gupta, R. K., and A. Przekwas. “Mathematical Models of Blast-Induced TBI: Current Status, Challenges, and Prospects.” Frontiers in Neurology, vol. 4, pp. 59, 2013.
- Traumatic Brain Injury Task Force. “Report to the Surgeon General.” 2007.
- Langlois, J. A., W. Rutland-Brown, and K. E. Thomas. Traumatic Brain Injury in the United States: Emergency Department Visits, Hospitalizations, and Deaths. Centers for Disease Control and Prevention, National Center for Injury Prevention and Control, Atlanta, GA, 2006.
- Defense and Veterans Brain Injury Center. http://www.dbvic.org/clinicalresearch.html, accessed November 2014.
- Taber, K., D. Warden, and R. Hurley. “Blast-Related Traumatic Brain Injury: What Is Known?” The Journal of Neuropsychiatry and Clinical Neurosciences, vol. 18, no. 2, pp. 141–145, 2006.
- Christman, C. W., M. S. Grady, S. A. Walker, K. L. Holloway, and J. T. Povlishock. “Ultrastructural Studies of Diffuse Axonal Injury in Humans.” Journal of Neurotrauma, vol. 11, no. 2, pp. 173–86, 1994.
- Smith, D. H., D. F. Meaney, and W. H. Shull. “Diffuse Axonal Injury in Head Trauma.” Journal of Head Trauma Rehabilitation, vol. 18, no. 4, pp. 307–316, 2003.
- Vettel, J. M., D. Bassett, R. Kraft, and S. Grafton. “Physics-Based Models of Brain Structure Connectivity Informed by Diffusion-Weighted Imaging.” ARL-RP-0355, U.S. Army Research Laboratory, Aberdeen Proving Ground, MD, 2010.
- Zhang L, K. H. Yang, and A. I. King. “A Proposed Injury Threshold for Mild Traumatic Brain Injury.” Journal of Biomechanical Engineering, vol.126, pp. 226–236, 2004.
- Wright, R.M., and K. T. Ramesh. “An Axonal Strain Injury Criterion for Traumatic Brain Injury.” Biomechics and Modeling in Mechanobiology, vol. 11, pp. 245–60, 2011.
- Bain, A.C., and D. F. Meaney. “Tissue-Level Thresholds for Axonal Damage in an Experimental Model of Central Nervous System White Matter Injury.” Journal of Biomechanical Engineering, vol. 122, pp. 615–622, 2000.
- Mendis, K. “Finite Element Modeling of the Brain to Establish Diffuse Axonal Injury Criteria.” Ph.D. thesis, Ohio State University, 1992.
- Ning, X., Q. Zhu, Y. Lanir, and S. S. Margulies. “A Transversely Isotropic Viscoelastic Constitutive Equation for Brainstem Undergoing Finite Deformation.” Journal of Biomechanical Engineering, vol. 128, pp. 925–930, 2006.
- Arbogast, K. B., and S. S. Margulies. “A Fiber-Reinforced Composite Model of the Viscoelastic Behavior of the Brainstem in Shear.” Journal of Biomechanics, vol. 32, pp. 865–870, 1999.
- Holzapfel, G. A. Nonlinear Solid Mechanics: A Continuum Approach for Engineering. John Wiley and Sons, LTD, 2000.
- Weiss, J. A., B. N. Maker, and S. Govindjee. “Finite Element Implementation of Incompressible, Transversely Isotropic Hyperelasticity.” Computer Methods in Applied Mechanics and Engineering, vol. 135, pp. 107–128, 1996.
- Kraft, R. H., and K. Ziegler. “High Rate Computational Brain Injury Biomechanics.” Proceedings of the ARL Ballistic Technology Workshop, 2010.
- Kraft, R., P. J. McKee, A. M. Dagro, and S. Grafton. “Combining the Finite Element Method With Structural Connectome-Based Analysis for Modeling Neurotrauma: Connectome Neurotrauma Mechanics.” PLOS Computational Biology, vol. 8, no. 8, 2012.
- Margulies, S. S., and L. E. Thibault. “A Proposed Tolerance Criterion for Diffuse Axonal Injury in Man.” Journal of Biomechanics, vol. 25, no. 8, pp. 917–23, 1992.
- Kleiven, S. “Predictors for Traumatic Brain Injuries Evaluated Through Accident Reconstructions.” Stapp Car Crash Journal, 2007.
- Bass, C. R., M. B. Panzer, K. A. Rafaels, G. Wood, J. Shridharani, and B. Capehart. “Brain Injuries from Blast.” Annals of Biomedical Engineering, vol. 40, no. 1, pp. 185–202, 2012.
- Dagro, A.; P. McKee, R. Kraft, and T. A. Zhang. “A Preliminary Investigation of Traumatically Induced Axonal Injury in a Three-Dimensional (3-D) Finite Element Model (FEM) of the Human Head During Blast-Loading.” ARL-TR-6504, U.S. Army Research Laboratory, Aberdeen Proving Ground, MD, 2013.
- McKee, P. J., A. M. Dagro, M. Vindiola, and J. M. Vettel. “Fiber Segment–Based Degradation Methods for a Finite Element-Informed Structural Brain Network.” ARL-TR-6739, U.S. Army Research Laboratory, Aberdeen Proving Ground, MD, 2013.